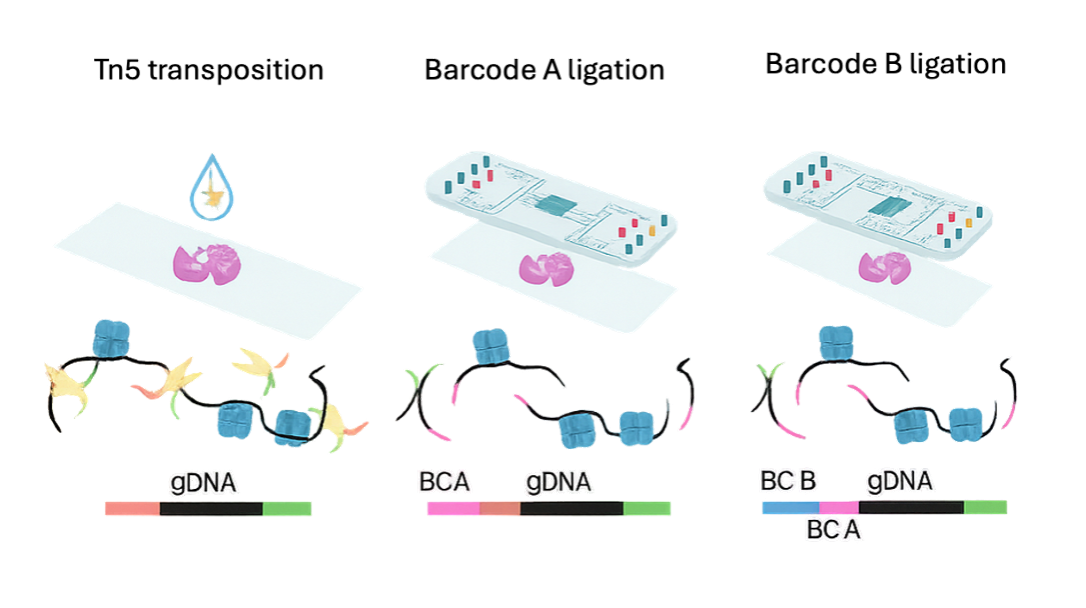
Spatial ATAC-seq
Uncover Chromatin Accessibility with Spatial Precision
Spatial ATAC-seq for high-resolution chromatin accessibility
Map open chromatin directly on tissue, connect accessibility with morphology, and uncover regulatory programs—on the same slide you already know and trust.
- Whole-tissue context
Preserve spatial organization while profiling chromatin accessibility across sections—no dissociation required. - From peaks to programs
Call peaks, quantify gene activity, score motifs/TF binding potential, and annotate neighborhoods & cell states. - Seamless co-analysis
Designed to integrate with spatial RNA or H&E for multi-omic interpretation and better target discovery. - FFPE support (new)
Run challenging archival samples with an FFPE-ready workflow and analysis presets.

New • FFPE support
FFPE-ready Spatial ATAC-seq
Unlock chromatin accessibility from archival tissue blocks. Preserve histology, maintain spatial context, and expand cohorts when fresh-frozen isn’t available.
- Archival reach: Compatible with FFPE sections to unlock retrospective studies and rare samples.
- Streamlined workflow: Slide-based chemistry with lab-friendly steps and QC checkpoints.
- Robust analysis: Gene activity, motif scoring, and peak calling presets tuned for FFPE data.
- Co-interpretation: Overlay with H&E or spatial RNA for richer biological context.
*Validated on representative oncology and immunology tissues. Contact us for sample guidelines.
Featured Spatial ATAC-seq Papers
Recent studies showcasing spatial chromatin accessibility across cancer, neurodegeneration, and human sensory tissue.

Spatial epigenomic niches underlie glioblastoma cell state plasticity
Reports spatially nested tumor niches in IDH-wildtype GBM and shows how local & long-range signals align
with chromatin accessibility states to drive plasticity.

Spatial profiling of chromatin accessibility reveals alteration of glial cells in Alzheimer’s disease mouse brain
Uses spatial ATAC-seq in 5xFAD versus control to chart accessibility shifts in microglia/other glia
and link motif programs to AD-related changes.

Epigenomic landscape of human dorsal root ganglion: sex differences and transcriptional regulation of nociceptive genes
Combines bulk & spatial ATAC-seq to reveal sex-linked accessibility differences in human DRG,
including X-chromosome–enriched DARs in females.
Contact Us
Have questions or interested in collaborating? We’d love to hear from you.
Get in Touch ↗